Nanoscale Mechanics
The interactions between proteins govern the interactions between and within cells. A critically important challenge in biophysics is to understand hydrophobic hydration, which describes the thermodynamic effects of adding oily species to water. Theoretical breakthroughs over recent years describe measurements that could solve this challenge. A major difficulty to overcome has been that hydrophobic hydration is an indirect effect, wherein much of the energy gained from two hydrophobic proteins interacting comes from favorable interactions among water molecules that are not forced to reside near oily surfaces. In a nutshell, the key theoretical advance has been the detailed prediction that the hydration energy depends on the length scale of the oily region. It depends on the shape and size of the cavity opened within the water to accommodate the oily species, and the energy scaling changes from cavity volume to cavity surface area when the radius is approximately 1 nm across. Our research group has made the key experimental measurements that tested and confirmed this prediction for synthetic polymers. Now is our first opportunity to test this model properly in proteins. Proteins are more complicated, of course: they change their structures when associating, and the energetic penalties for association depend on the oil–water interactions that are removed, which are typically due to local oily patches on the protein rather than the whole. What is needed is a model that can accommodate both local patch and whole protein length scales and, just as importantly, experimental data to test the model. Fortunately, there exist new structural data for key protein aggregates of amyloid beta (Aβ), whose toxicity in Alzheimer’s disease is often linked to its misfolding and aggregation. We aim to combine these structural data with new data on hydrophobic hydration energetics. This could lay the groundwork for a method to screen for ligands that can interfere with the development of toxic aggregates.
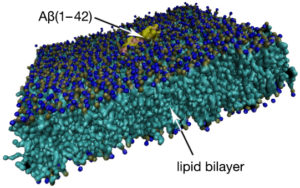
Simulation of Aβ1–42 monomer (PDB: 1IYT) at a lipid bilayer using a Martini forcefield. The membrane has a surface area of 15 × 15 nm² and contains 1,2-dipalmitoyl-sn-glycero-3-phosphocholine, cholesterol, and monosialotetrahexo-sylganglioside at a 65:30:5 ratio.
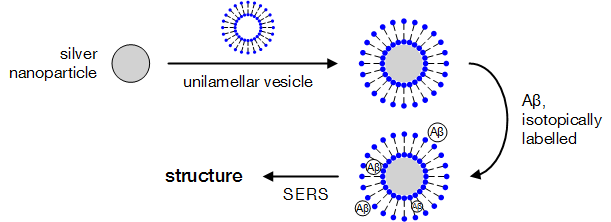
Phospholipid-encapsulated silver nanoparticles may be incubated with Aβ oligomers, leading to complexes suitable for structural studies via surface-enhanced Raman spectroscopy.
Animation displaying how a force curve is generated from a single-polymer pulling experiment.
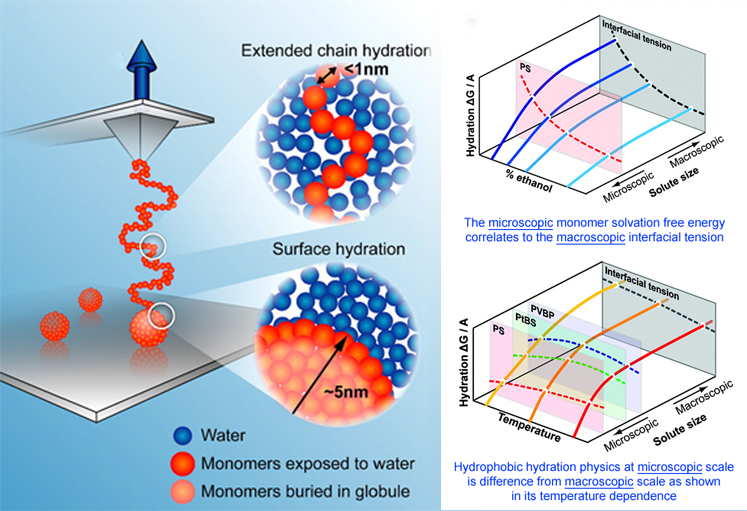
The extension of a single, hydrophobic polymer drives a globule-to-coil structural transition. The relative hydration length scales associated with the collapsed globule and extended coil are shown in the insets.
Representative publications
Mondal, J.; Halverson, D.; Li, I. T. S.; Stirnemann, G.; Walker, G. C.; Berne, B. J. How osmolytes influence hydrophobic polymer conformations: A unified view from experiment and theory. Proc. Natl. Acad. Sci. U. S. A. 2015, 112, 9270–9275. doi: 10.1073/pnas.1511780112.
Li, I. T. S.; Walker, G. C. Single Polymer Studies of Hydrophobic Hydration. Acc. Chem. Res. 2012, 45, 2011–2021. doi: 10.1021/ar200285h.
Li, I. T. S.; Walker, G. C. Signature of hydrophobic hydration in a single polymer. Proc. Natl. Acad. Sci. U. S. A. 2011, 108, 16527–16532. doi: 10.1073/pnas.1105450108.
Li, I. T. S.; Walker, G. C. Interfacial Free Energy Governs Single Polystyrene Chain Collapse in Water and Aqueous Solutions. J. Am. Chem. Soc. 2010, 132, 6530–6540. doi: 10.1021/ja101155h.
